What'sNEW August–September 2008
 30 September 2008 30 September 2008
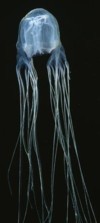 Horizontal gene transfer with the animals is going to turn out to be more widespread than anybody believes now. When that realization comes down, it will definitely change the way people think about evolution. — Michael Syvanen, Professor of Medical Microbiology & Immunology, School of Medicine, University of California at Davis
Horizontal gene transfer with the animals is going to turn out to be more widespread than anybody believes now. When that realization comes down, it will definitely change the way people think about evolution. — Michael Syvanen, Professor of Medical Microbiology & Immunology, School of Medicine, University of California at DavisSyvanen is quoted in Nature's commentary on another example of trans-kingdom horizontal gene transfer (HGT), this one observed by team of evolutionary biologists at the Pierre and Marie Curie University in Paris. They found evidence that an ancestor of cnidarians (sea anemones, jellyfish, corals etc.) acquired an essential gene from bacteria. The gene encodes a subunit of PGA synthease, enabling cnidarians to sting their prey. "The result was a great surprise," says team leader Nicolas Rabet. In cosmic ancestry such results are expected.
 Elsa Denker et al., "Horizontal gene transfer and the evolution of cnidarian stinging cells" [abstract], p R858-R859 v 18, Current Biology, 23 Sep 2008. Elsa Denker et al., "Horizontal gene transfer and the evolution of cnidarian stinging cells" [abstract], p R858-R859 v 18, Current Biology, 23 Sep 2008.
 Amber Dance, "How the jellyfish got its sting" doi:10.1038/news.2008.1134, [html], Nature, online 28 Sep 2008. Amber Dance, "How the jellyfish got its sting" doi:10.1038/news.2008.1134, [html], Nature, online 28 Sep 2008.
 Viruses and Other Gene Transfer Mechanisms is the main CA webpage about HGT Viruses and Other Gene Transfer Mechanisms is the main CA webpage about HGT
[ What'sNEW about HGT What'sNEW about HGT  ] ]
 Horizontal Gene Transfer...: we briefly review this book edited by Michael Syvanen and Clarence Kato, 4 Nov 1999. Horizontal Gene Transfer...: we briefly review this book edited by Michael Syvanen and Clarence Kato, 4 Nov 1999.
 20 September 2008 20 September 2008
 Woodstock of evolution? That's how Nature descibes a meeting among sixteen leading biologists at the Konrad Lorenz Institute for Evolution and Cognition Research in Altenberg, Austria, 10-13 July 2008. The purpose of the meeting was to go beyond the modern synthesis that has held evolutionary theory together for more than sixty years. This is necessary because the existing theory leaves much unexplained —
Woodstock of evolution? That's how Nature descibes a meeting among sixteen leading biologists at the Konrad Lorenz Institute for Evolution and Cognition Research in Altenberg, Austria, 10-13 July 2008. The purpose of the meeting was to go beyond the modern synthesis that has held evolutionary theory together for more than sixty years. This is necessary because the existing theory leaves much unexplained —
- "When the public thinks about evolution, they think about the origin of wings and the invasion of the land," says Graham Budd, a palaeobiologist at the University of Uppsala, Sweden. "But these are things that evolutionary theory has told us little about."
- "The modern synthesis is remarkably good at modelling the survival of the fittest, but not good at modelling the arrival of the fittest," comments Scott Gilbert, an evo-devo researcher at Swarthmore College in Pennsylvania.
The participants agreed that such shortcomongs must be carefully downplayed (see right), because "creationists seize on any hint of splits in evolutionary theory or dissatisfaction with Darwinism." Apparently darwinism is still gridlocked and real reform is not imminent. Meanwhile, we noticed a couple of reported comments that are consistent with cosmic ancestry —
- Stuart Newman, a developmental biologist at New York Medical College, comments "You can't deny the force of selection in genetic evolution, but in my view this is stabilizing and fine-tuning forms that originate due to other processes."
- To explain the sudden appearance of turtles, Scott Gilbert suggests, "all the genes are probably there already...."
 John Whitfield, "Postmodern evolution?" [html], doi:10.1038/455281a, p 281-284 v 455, Nature, 18 Sep 2008. John Whitfield, "Postmodern evolution?" [html], doi:10.1038/455281a, p 281-284 v 455, Nature, 18 Sep 2008.
 Elizabeth Pennisi, "Modernizing the Modern Synthesis" [summary], doi:10.1126/science.321.5886.196, p 196-197 v 321, Science, 11 Jul 2008. Elizabeth Pennisi, "Modernizing the Modern Synthesis" [summary], doi:10.1126/science.321.5886.196, p 196-197 v 321, Science, 11 Jul 2008.
 Toward an Extended Evolutionary Synthesis: 18th Altenberg Workshop in Theoretical Biology 2008, organized by Massimo Pigliucci and Gerd B. Müller. Toward an Extended Evolutionary Synthesis: 18th Altenberg Workshop in Theoretical Biology 2008, organized by Massimo Pigliucci and Gerd B. Müller.
 Altenberg! The Woodstock of Evolution? by Suzan Mazur, Scoop, 4 Mar 2008. Altenberg! The Woodstock of Evolution? by Suzan Mazur, Scoop, 4 Mar 2008.
 Altenberg 16: An Exposé Of The Evolution Industry by Suzan Mazur, Scoop, 6 Jul 2008. Altenberg 16: An Exposé Of The Evolution Industry by Suzan Mazur, Scoop, 6 Jul 2008.
 Does Microevolution Explain Macroevolution?, a section of CA's webpage about neodarwinism, considers "stabilizing and fine-tuning" versus "forms that originate due to other processes." Does Microevolution Explain Macroevolution?, a section of CA's webpage about neodarwinism, considers "stabilizing and fine-tuning" versus "forms that originate due to other processes."
 Macroevolutionary Progress Redefined... is another CA webpage about the difference between micro- and macroevolution. Macroevolutionary Progress Redefined... is another CA webpage about the difference between micro- and macroevolution.
 Metazoan Genes Older Than Metazoa? is a CA webpage with many examples of genes that would be "there already." Metazoan Genes Older Than Metazoa? is a CA webpage with many examples of genes that would be "there already."
 Evolution versus Creationism is a CA webpage that discusses "gridlock." Evolution versus Creationism is a CA webpage that discusses "gridlock."
 The World Summit on Evolution, another meeting described as "The Woodstock of Evolution," What'sNEW, 14 Jul 2005. The World Summit on Evolution, another meeting described as "The Woodstock of Evolution," What'sNEW, 14 Jul 2005.
 Suzan Mazur sends a link to her interview with Stuart Newman, "The Origin of Form Was Abrupt...," 10 Oct 2008. Suzan Mazur sends a link to her interview with Stuart Newman, "The Origin of Form Was Abrupt...," 10 Oct 2008.

 18 September 2008 18 September 2008
The virophage could be a vehicle mediating lateral gene transfer between giant viruses.
 Bernard La Scola, Christelle Desnues et al., "The virophage as a unique parasite of the giant mimivirus" [abstract], doi:10.1038/nature07218, p 100-104 v 455, Nature, 4 Sep 2008. Bernard La Scola, Christelle Desnues et al., "The virophage as a unique parasite of the giant mimivirus" [abstract], doi:10.1038/nature07218, p 100-104 v 455, Nature, 4 Sep 2008.
 Hiroyuki Ogata and Jean-Michel Claverie, "How to Infect a Mimivirus" [link], doi:10.1126/science.1164839, p 1305-1306 v 321, Science, 5 Sep 2008. Hiroyuki Ogata and Jean-Michel Claverie, "How to Infect a Mimivirus" [link], doi:10.1126/science.1164839, p 1305-1306 v 321, Science, 5 Sep 2008.
 Viruses... is the main CA webpage about HGT Viruses... is the main CA webpage about HGT
[ What'sNEW about HGT What'sNEW about HGT  ] ]
|

Putative functions of 37 genes acquired from non-organellar sources prior to the split of red algae and green plants. |
 11 September 2008 11 September 2008
...Multiple independently acquired genes are able to generate and optimize key evolutionary novelties in major eukaryotic groups. This conclusion comes from an extensive genomic analysis of an extremophile red alga, Cyanidioschyzon. The two analysts acknowledge that genes may be transferred from organellar genomes or by hybridization, but they exclusively consider gene transfer by other means. Biologists Jinling Huang and Peter Gogarten observe —
- Some anciently acquired novel genes identified in our analyses appear to be critical for plant development or adaptation.
- An interesting observation from the studies of HGT in eukaryotes is that the vast majority of well-documented cases involve prokaryotes as donors.
- It is interesting to speculate whether early eukaryotes continuously obtained genes from a larger prokaryotic gene pool, either individually or occasionally in large chunks....
- The multiple introductions of the same gene from various prokaryotic sources ...suggest that HGT is a continuous and dynamic process.
- The barriers to interdomain gene transfer during early eukaryotic evolution might not [have been] as significant as observed today.
- By acquiring ready-to-use genes from other sources, HGT avoids a slow process of gene generation and might confer to the recipient organisms immediate abilities to explore new resources and niches.
- It is possible that the identified genes in our analyses represent only the tip of an iceberg for the overall scope of ancient HGT in eukaryotes.
The importance of horizontal gene transfer (HGT) in eukaryotic evolution is becoming better recognized. Meanwhile, the present study admits, HGT already "has revolutionized our view of microbial evolution." These developments confirm a basic prediction of cosmic ancestry, in which HGT is essential for macroevolutionary progress in all domains of life. For strict darwinism, HGT is a surprise no longer easy to ignore.
 Jinling Huang and J Peter Gogarten, "Concerted gene recruitment in early plant evolution" [abstract], doi:10.1186/gb-2008-9-7-r109, 9:R109, Genome Biology, 8 Jul 2008. Jinling Huang and J Peter Gogarten, "Concerted gene recruitment in early plant evolution" [abstract], doi:10.1186/gb-2008-9-7-r109, 9:R109, Genome Biology, 8 Jul 2008.
 Viruses and Other Gene Transfer Mechanisms is the main CA webpage about HGT Viruses and Other Gene Transfer Mechanisms is the main CA webpage about HGT
[ What'sNEW about HGT What'sNEW about HGT  ] ]
 Thanks, everyone. Thanks, everyone.
 10 September 2008 10 September 2008
Tardigrades survived ten days in space on the European Space Agency's Foton M3 mission last September. The vacuum and dryness were well-tolerated by the two species tested, but radiation was lethal to some. Also known as waterbears, tardigrades range in size from 1.5 to .05 millimeter. "This proves that at least some animals can survive the rigors of space flight unprotected." K. Ingemar Jönsson of Sweden's Kristianstad University led the team that designed the experiment.

 K. Ingemar Jönsson et al., "Tardigrades survive exposure to space in low Earth orbit" [summary], doi:10.1016/j.cub.2008.06.048, p R729-R731 v 18, Current Biology, 9 Sep 2008. K. Ingemar Jönsson et al., "Tardigrades survive exposure to space in low Earth orbit" [summary], doi:10.1016/j.cub.2008.06.048, p R729-R731 v 18, Current Biology, 9 Sep 2008.
 Tardigrades from Sweden first animal in open space conditions, Kristianstad University, 21 Sep 2007. Tardigrades from Sweden first animal in open space conditions, Kristianstad University, 21 Sep 2007.
 Space Suits Them: First Animal That Can Survive in Orbit, by David Biello, Scientific American, 8 Sep 2008. Space Suits Them: First Animal That Can Survive in Orbit, by David Biello, Scientific American, 8 Sep 2008.
 Creature Survives Naked in Space, Space.com, 8 Sep 2008. Creature Survives Naked in Space, Space.com, 8 Sep 2008.
 Tiny 'water bears' can survive in outer space, The Nation, 11 Sep 2008. Tiny 'water bears' can survive in outer space, The Nation, 11 Sep 2008.
 Eight-Legged Space Survivor Gives 'Panspermia' New Life by Robert Roy Britt, Space.com, 16 Sep 2008. Eight-Legged Space Survivor Gives 'Panspermia' New Life by Robert Roy Britt, Space.com, 16 Sep 2008.
 'Alien' Water Bears Amaze Scientists by Lee Pullen, Astrobiology Magazine, 16 Oct 2008. 'Alien' Water Bears Amaze Scientists by Lee Pullen, Astrobiology Magazine, 16 Oct 2008.
 Introduction: More than Panspermia is a related CA webpage. Introduction: More than Panspermia is a related CA webpage.
 Thanks, Cody Bennett, Jim Galasyn, Ken Augustyn, Bill and Ann Tucker, Chandra Wickramasinghe and Google Alerts. Thanks, Cody Bennett, Jim Galasyn, Ken Augustyn, Bill and Ann Tucker, Chandra Wickramasinghe and Google Alerts.
 26 August 2008 26 August 2008
Proofs That Life Is Cosmic, by Fred Hoyle and Chandra Wickramasinghe, can now be downloaded in full for free from the Buckingham Centre for Astrobiology (BCAB) and from this website. The PDF pages are scanned images. Although we are long familiar with this work, we are still impressed with the farseeing insights it contains. We may host other early works by these authors as they become available.
 Fred Hoyle and Chandra Wickramasinghe, Proofs that Life is Cosmic, n 1, Memoirs of the Institute of Fundamental Studies, Sri Lanka, Dec 1982. Fred Hoyle and Chandra Wickramasinghe, Proofs that Life is Cosmic, n 1, Memoirs of the Institute of Fundamental Studies, Sri Lanka, Dec 1982.
 Hoyle and Wickramasinghe's Analysis of Interstellar Dust is a related CA webpage with a list of Selected Resources. Hoyle and Wickramasinghe's Analysis of Interstellar Dust is a related CA webpage with a list of Selected Resources.
 Three more early works by Hoyle and Wickramasinghe are available for download, 9 Oct 2008. Three more early works by Hoyle and Wickramasinghe are available for download, 9 Oct 2008.
 22 August 2008 22 August 2008
It is now completely clear that genomic complexity was present very early on — Casey Dunn, Brown University
An international biology team has analysized the genome of a tiny flat marine organism, Trichoplax adhaerens. The species is known to have only four cell types, no head or tail, no stomach, no nerves and no muscles. It reproduces asexually (without meoisis), by fission. Earlier studies of mitichondrial DNA indicated that its phylum, Placozoa, may be the "earliest branching basal metazoan phylum." The new analysis is surprising.
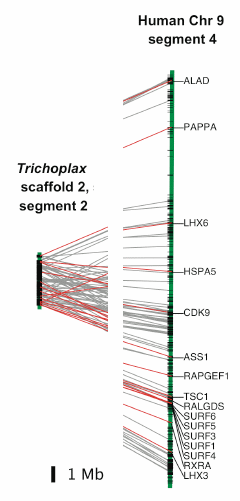 The Trichoplax adhaerens genome contains 98 million base pairs, and an estimated 11,514 protein-coding genes. Unlike other more advanced animals such as fruitflies and nematodes, Trichoplax genes have an intron density (7.6/kb) similar to that of vertebrates (8.5/kb). Many of these introns (82% in conserved regions) occupy the same position and phase as the corresponding ones in humans. Even more interestingly, "Each of the 21 longest gene-rich Trichoplax scaffolds contains segments with a significant concentration of orthologues on one or more human chromosome segments." For example, the illustrated Trichoplax and human segments contain 83 pairs of orthologs, of which the "well-known" ones are connected with red or gray lines (disconnected in our adaptation of this image.)
The Trichoplax adhaerens genome contains 98 million base pairs, and an estimated 11,514 protein-coding genes. Unlike other more advanced animals such as fruitflies and nematodes, Trichoplax genes have an intron density (7.6/kb) similar to that of vertebrates (8.5/kb). Many of these introns (82% in conserved regions) occupy the same position and phase as the corresponding ones in humans. Even more interestingly, "Each of the 21 longest gene-rich Trichoplax scaffolds contains segments with a significant concentration of orthologues on one or more human chromosome segments." For example, the illustrated Trichoplax and human segments contain 83 pairs of orthologs, of which the "well-known" ones are connected with red or gray lines (disconnected in our adaptation of this image.)
Most surprisingly, many Trichoplax genes with orthologs in more advanced species code for complex features that Trichoplax itself does not have. For example, the analysis reports:
- "Trichoplax contains a rich repertoire of transcription factors ...commonly associated with patterning and regionalization during eumetazoan development....
- "Transcription factors that regulate cell type specification and differentiation in bilaterians are also abundant in Trichoplax....
- "Trichoplax ... possesses ...no evident homology to bilaterian anteroposterior or dorsoventral axes. Yet, components of a complete ...signalling pathway—used for axial patterning in bilaterians and cnidarians and in demosponge larvae—are present....
- "Components of other signalling pathways such as the Notch and JAK/STAT pathways are present, but these pathways seem to be incomplete....
- "...We find various ion channels that are implicated in neural signalling in animals.
- "Components of neurotransmitter biosynthesis and vesicle transport systems, as well as a putative neuroendocrine-like secretory apparatus, are also found in the genome.
- "Putative neurotransmitter and neuropeptide receptors are also present....
- "Synapse formation proteins ...and structural elements of the post-synaptic scaffold known to be present in sponges ...are also found in Trichoplax, including the receptors and channels missing from the sponge genome.
- "Genes associated with neural migration and axon guidance in bilaterians ...are also present.
- "Trichoplax ...is reported to lack ...any described extracellular matrix (ECM). Its genome, however, contains a diverse set of genes that code for putative ECM proteins. These include collagen IV, laminin-alpha, -beta and -gamma, and nidogen; however, fibronectin, fibrin, elastin and vitronectin are all apparently absent.
- "The Trichoplax genome also encodes cell-surface adhesion proteins ...that interact with each other and the ECM in bilaterians, and encodes cytoskeletal linker proteins ...that help organize the actin cytoskeleton and/or transduce signals in other eumetazoans.
- "Similarly, protein components ...that would permit dual functions of beta-catenin and integrin receptors in adhesion and signal transduction ...are encoded by the genome.
- "Enzyme families known to modify ECM components and/or signalling molecules in the matrix ...are also present.
- "...Meiosis-associated genes are found in the genome.
- "Although the Trichoplax body plan is simple, its genome encodes a rich array of transcription factors and signalling pathways that are typically associated with eumetazoan developmental patterning and cell-type specification. A question remains: what role do these genes have in placozoans?"

Mansi Srivastava | |
Under darwinian theory, natural selection is essential for the composition of genes for new functions. Of course, natural selection can only operate on functions when they are expressed. But apparently, these functions did not begin to be expressed until long after the genes encoding them were extant. Under darwinian theory, something is badly wrong here.The findings support a basic prediction of cosmic ancestry, namely that all genes are very old.
 Mansi Srivastava et al., "The Trichoplax genome and the nature of placozoans" [abstract], doi:10.1038/nature07191, p 955-960 v 454, Nature, 21 Aug 2008. Mansi Srivastava et al., "The Trichoplax genome and the nature of placozoans" [abstract], doi:10.1038/nature07191, p 955-960 v 454, Nature, 21 Aug 2008.
 Mansi Srivastava interviewed [html], doi:10.1038/7207xib, p xi v 454, Nature, 21 Aug 2008. Mansi Srivastava interviewed [html], doi:10.1038/7207xib, p xi v 454, Nature, 21 Aug 2008.
 Elizabeth Pennisi, "'Simple' Animal's Genome Proves Unexpectedly Complex" [summary], p 1028-1029 v 321, Science, 22 Aug 2008. Elizabeth Pennisi, "'Simple' Animal's Genome Proves Unexpectedly Complex" [summary], p 1028-1029 v 321, Science, 22 Aug 2008.
 The Dunn Lab, Brown University, Casey Dunn, Principal Investigator. The Dunn Lab, Brown University, Casey Dunn, Principal Investigator.
 Metazoan Genes Older Than Metazoa? is a related CA webpage with links to examples like the mentioned sponges, also discussed yesterday. Metazoan Genes Older Than Metazoa? is a related CA webpage with links to examples like the mentioned sponges, also discussed yesterday.
 New genetic programs in Darwinism and strong panspermia is a related CA webpage. New genetic programs in Darwinism and strong panspermia is a related CA webpage.
 21 August 2008 21 August 2008
 Sponges don't have nerve cells, yet they have genes for directing the formation of nerves. "What was really cool is we took some of these genes and expressed them in frog and flies and the sponge gene became functional – the sponge gene directed the formation of nerves in these more complex animals," says Professor Bernie Degnan (pictured) of the School of Integrative Biology, University of Queensland. His lab is mapping the entire genome of the sea sponge. "We really didn't expect it," he adds.
Sponges don't have nerve cells, yet they have genes for directing the formation of nerves. "What was really cool is we took some of these genes and expressed them in frog and flies and the sponge gene became functional – the sponge gene directed the formation of nerves in these more complex animals," says Professor Bernie Degnan (pictured) of the School of Integrative Biology, University of Queensland. His lab is mapping the entire genome of the sea sponge. "We really didn't expect it," he adds. The data reveal nothing about the origin of these genes, only that they must be older than previously thought. Following darwinian logic the research team adopts a startling conclusion — "The bilaterian neurogenic circuit... was functional in the very first metazoans." The data are not surprising for cosmic ancestry, in which genes must be available before they can be expressed. If so, sometimes they would reside in species with no use for them, as seems to be the case here.
 Gemma S. Richards, Elena Simionato et al., "Sponge Genes Provide New Insight into the Evolutionary Origin of the Neurogenic Circuit" [abstract], p 1156-1161 v 18, Current Biology, 05 Aug 2008. Gemma S. Richards, Elena Simionato et al., "Sponge Genes Provide New Insight into the Evolutionary Origin of the Neurogenic Circuit" [abstract], p 1156-1161 v 18, Current Biology, 05 Aug 2008.
 UQ research touches a nerve, University of Queensland, 20 Aug 2008. UQ research touches a nerve, University of Queensland, 20 Aug 2008.
 Metazoan Genes Older Than Metazoa? is a related CA webpage. Metazoan Genes Older Than Metazoa? is a related CA webpage.
 Thanks, Hans-Peter Wheeler. Thanks, Hans-Peter Wheeler.
 17 August 2008 17 August 2008
The origin of new genes in well-documented species of fruitflies is the focus of an exhaustive study by geneticists of the Kunming Institute of Zoology, Chinese Academy of Sciences. They conclude that 5 to 11 functional new genes per million years originate in the D. melanogaster subgroup. Of these new genes —
- Most arise by gene duplication, either tandem or dispersed. The dispersed ones (which may have originated as tandem ones) are more likely to be retained.
- 10.2% come from retroposition, the generation of an intronless copy by reverse transcription of mRNA. (This percentage would likely be higher among species with more active retroposons, such as humans and rice.)
- Surprisingly, 11.9% originate from previously noncoding sequences. They call these de novo genes.
- "Chimeric structures significantly contribute to the evolution of new genes."
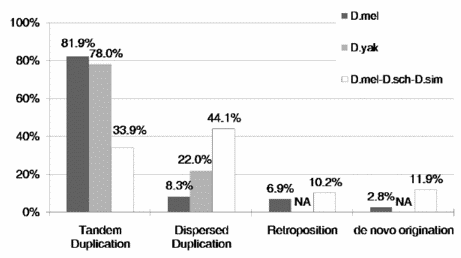 We are very interested in the findings of this study. We specifically want to know if darwinian processes can create new genetic instructions for new functions (new encoded meaning), as is generally assumed. This study does not consider any specific functions, new or not, that the new genes may encode. But with lengthy ORFs and clear evidence of functional constraint, the genes must be doing something. The ones arising from previously noncoding sequences obviously provide something not already expressed in fruitflies. But do the new genes create new meaning?
We are very interested in the findings of this study. We specifically want to know if darwinian processes can create new genetic instructions for new functions (new encoded meaning), as is generally assumed. This study does not consider any specific functions, new or not, that the new genes may encode. But with lengthy ORFs and clear evidence of functional constraint, the genes must be doing something. The ones arising from previously noncoding sequences obviously provide something not already expressed in fruitflies. But do the new genes create new meaning?
Other evidence shows that duplication usually allows previously existing functions to segregate and optimize. And retroposition, by itself, creates no new meaning. But duplicates and retroposed genes may acquire extraneous protein-coding or regulatory sequences, and novel phenotypes may emerge. We suggest that these chimeric genes are created by processes similar to those in computer models where shuffling algorithms, like suffling exons, can create variety. But these process have not been shown to create new meaning. They only explore the potential of the encoded meaning that is already available.
The geneticists call new genes coming from previously noncoding sequences de novo. Their nine examples map to homologous noncoding regions found so far only in other insects. Most are present in a fruitfly outgroup, D. yakuba, but one is not — its nearest relative known to have a matching sequence is the honeybee (implicating horizontal gene transfer). And they observe, "Notably, the lengths of all the newly identified de novo genes' protein products are predicted to be longer than 100 amino acids." Of course, a random DNA sequence longer than 300 nucleotides (100 codons) is absurdly unlikely to code for a useful protein, yet these de novo sequences apparently do. They must be not random sequences. Instead they appear to be pre-existing genes not previously expressed in any sequenced species.
In our proposed amendment to darwinian theory, evolutionary advances occur when genetic programs are acquired, reassembled, activated and optimised. This scenario is plausible and can be demonstrated with computer models. We think the evidence in the present study supports this scenario. Contrarily, in standard darwinian theory new genetic programs must be written originally from scratch. In our opinion, nothing in the present study even hints at such a process.
| In a strict sense, nothing in evolution is generated de novo — Susumo Ohno, 1970 |
 Qi Zhou, Guojie Zhang, Yue Zhang et al., ""On the origin of new genes in Drosophila" [abstract], doi:10.1101/gr.076588.108, p 1446-1455 v 18, Genome Research, Sep (online 11 Jun) 2008. Qi Zhou, Guojie Zhang, Yue Zhang et al., ""On the origin of new genes in Drosophila" [abstract], doi:10.1101/gr.076588.108, p 1446-1455 v 18, Genome Research, Sep (online 11 Jun) 2008.
 Viruses and Other Gene Transfer Mechanisms is the main CA webpage about gene acquisition Viruses and Other Gene Transfer Mechanisms is the main CA webpage about gene acquisition
[ What'sNEW about HGT What'sNEW about HGT  ] ]
 Testing Darwinism versus Cosmic Ancestry is a related CA webpage. Testing Darwinism versus Cosmic Ancestry is a related CA webpage.
 Thanks, Wen Wang and many others. Thanks, Wen Wang and many others.
 13 Sep 2010: Origins, evolution, and phenotypic impact of new genes cites the research discussed here. 13 Sep 2010: Origins, evolution, and phenotypic impact of new genes cites the research discussed here.
 8 August 2008 8 August 2008
Something is missing in our understanding of how evolution produced complex creatures. I don't know what it is, nor do I think anyone else does, contrary to the claims you hear asserted, says Mark Bedau (pictured), co-organizer of the Eleventh International Conference on the Simulation and Synthesis of Living Systems. Mark has advised us in our own efforts to explore the range of evolution in closed or quarantined systems, but this admission is news to us. Bedau also remarks, Unlike the real world, the outcome of computer evolution is built into its programming.
 But Mark, why is this unlike the real world? In our observation, biological evolution depends on its programming in the same way. New complex features readily emerge when the genetic programming for them is installed and activated by well-studied processes including horizontal gene transfer (HGT). For evolution among prokaryotes, this understanding is already in place, but the significance of this major paradigm shift goes unnoticed. For evolution among eukaryotes, the situation reminds us of that for prokaryotes a decade ago — examples of HGT with important evolutionary effects are accumulating rapidly.
But Mark, why is this unlike the real world? In our observation, biological evolution depends on its programming in the same way. New complex features readily emerge when the genetic programming for them is installed and activated by well-studied processes including horizontal gene transfer (HGT). For evolution among prokaryotes, this understanding is already in place, but the significance of this major paradigm shift goes unnoticed. For evolution among eukaryotes, the situation reminds us of that for prokaryotes a decade ago — examples of HGT with important evolutionary effects are accumulating rapidly.
We congratulate Mark for his straightforwardness. We hope he and others will seriously consider the possibility that the outcome of biological evolution is also built into its programming.
 Can we make software that comes to life? Telegraph.co.uk, 5 Aug 2008. Can we make software that comes to life? Telegraph.co.uk, 5 Aug 2008.
 Dave Hillis wonders if ALife is "moving away from Darwinism but not from the natural world after all," in email posted on Evolutionprize.net, 5 Aug 2008. Dave Hillis wonders if ALife is "moving away from Darwinism but not from the natural world after all," in email posted on Evolutionprize.net, 5 Aug 2008.
 Can Computers Mimic Darwinian Evolution? is one of several related CA webpages. Can Computers Mimic Darwinian Evolution? is one of several related CA webpages.
 The number of cases of HGT involving eukaryotes..., What'sNEW, 20 Jul 2008. The number of cases of HGT involving eukaryotes..., What'sNEW, 20 Jul 2008.
 Thanks, Stan Franklin. Thanks, Stan Franklin.
 Mark Bedau comments, 23 Sep 2008. Mark Bedau comments, 23 Sep 2008.
 7 August 2008 7 August 2008
Finding perchlorates is neither good nor bad for life, but it does make us reassess how we think about life on Mars — Michael Hecht of NASA's Jet Propulsion Laboratory
 Kenneth Chang, "A Finding, Perhaps, but Not of Mars Life" [online version], p A20, The New York Times, 6 Aug 2008. Kenneth Chang, "A Finding, Perhaps, but Not of Mars Life" [online version], p A20, The New York Times, 6 Aug 2008.
 Perchlorate found on Mars, doi:10.1038/news.2008.1016, by Eric Hand, NatureNews, 6 Aug 2008. Perchlorate found on Mars, doi:10.1038/news.2008.1016, by Eric Hand, NatureNews, 6 Aug 2008.
 Phoenix Mars Team Opens Window on Scientific Process, NASA/JPL, 5 Aug 2008. Phoenix Mars Team Opens Window on Scientific Process, NASA/JPL, 5 Aug 2008.
 3 August 2008 3 August 2008
 The potential for life on Mars at the Phoenix landing site is believed to be the subject of a forthcoming NASA announcement.
The potential for life on Mars at the Phoenix landing site is believed to be the subject of a forthcoming NASA announcement.
 White House Briefed On Potential For Mars Life, MarsToday.com, 1 Aug 2008. White House Briefed On Potential For Mars Life, MarsToday.com, 1 Aug 2008.
 The White House is Briefed: Phoenix About to Announce 'Potential For Life' on Mars, UniverseToday.com, 1 Aug 2008. The White House is Briefed: Phoenix About to Announce 'Potential For Life' on Mars, UniverseToday.com, 1 Aug 2008.
 Chandra Wickramasinghe Says there is Contemporary Life on the Red Planet Mars, by Walter Jayawardhana, Lankaweb, Sri Lanka, 5 Aug 2008. Chandra Wickramasinghe Says there is Contemporary Life on the Red Planet Mars, by Walter Jayawardhana, Lankaweb, Sri Lanka, 5 Aug 2008.
 Thanks, Jim Galasyn, Larry Klaes and Google Alerts. Thanks, Jim Galasyn, Larry Klaes and Google Alerts.
 2 August 2008 2 August 2008
This is the first time Martian water has been touched and tasted — William Boynton, lead scientist for the Thermal and Evolved-Gas Analyzer on the Phoenix Mars Lander that touched down 25 May.
 NASA Spacecraft Confirms Martian Water, Mission Extended, NASA / University of Arizona, 31 Jul 2008. NASA Spacecraft Confirms Martian Water, Mission Extended, NASA / University of Arizona, 31 Jul 2008.
 Breaking news: water on Mars, Nature.com, 1 Aug 2008. Breaking news: water on Mars, Nature.com, 1 Aug 2008.
 Life on Mars! is a related CA webpage. Life on Mars! is a related CA webpage.
 Thanks, Rob Cooper and Ellen Cooper Klyce. Thanks, Rob Cooper and Ellen Cooper Klyce.
 1 August 2008 1 August 2008
Our results indicate that, on average, at least 81 ± 15% of the genes in each genome studied were involved in lateral gene transfer at some point in their history, even though they can be vertically inherited after acquisition, uncovering a substantial cumulative effect of lateral gene transfer on longer evolutionary time scales. This result comes from geneticists in Düsseldorf and Tel Aviv using a new method of network analysis that reveals "both vertical and lateral components of evolutionary history among 539,723 genes distributed across 181 sequenced prokaryotic genomes." In their report they comment — - Once acquired, genes can be vertically inherited within a group, and the MLN [minimal lateral network] suggests that this has occurred for the vast majority of genes, and probably all, given that we have inferred no LGT [lateral gene transfer] events from conflicting gene trees, during prokaryote genome evolution.
- Our findings indicate that, over long evolutionary time scales, the cumulative role of LGT leaves almost no gene family among prokaryotes untouched.
- The conservative lower bound nature of our method for inferring LGT among prokaryotes indicates that evolution by lateral transfer affects the vast majority of gene families, and probably all, but possibly at a low rate.
If all genes in prokaryotes have probably been acquired by LGT — also called horizontal gene transfer (HGT) — the standard darwinian explanation for where genes come from is inadequate. We are astounded by the lack of attention to this crisis.
 Tal Dagan, Yael Artzy-Randrup and William Martin, "Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution" [Open Access abstract], doi:10.1073/pnas.0800679105, p 10039-10044 v 105, Proc. Natl. Acad. Sci. USA, 22 Jul (online 16 Jul) 2008. Tal Dagan, Yael Artzy-Randrup and William Martin, "Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution" [Open Access abstract], doi:10.1073/pnas.0800679105, p 10039-10044 v 105, Proc. Natl. Acad. Sci. USA, 22 Jul (online 16 Jul) 2008.
 Laura M. Zahn, "Taking the Long View" [html], doi:10.1126/science.321.5890.747b, p 747 v 321, Science, 8 Aug 2008. Laura M. Zahn, "Taking the Long View" [html], doi:10.1126/science.321.5890.747b, p 747 v 321, Science, 8 Aug 2008.
 Viruses and Other Gene Transfer Mechanisms is the main CA webpage about gene acquisition Viruses and Other Gene Transfer Mechanisms is the main CA webpage about gene acquisition
[ What'sNEW about HGT What'sNEW about HGT  ] ]
|
 Horizontal gene transfer with the animals is going to turn out to be more widespread than anybody believes now. When that realization comes down, it will definitely change the way people think about evolution. — Michael Syvanen, Professor of Medical Microbiology & Immunology, School of Medicine, University of California at Davis
Horizontal gene transfer with the animals is going to turn out to be more widespread than anybody believes now. When that realization comes down, it will definitely change the way people think about evolution. — Michael Syvanen, Professor of Medical Microbiology & Immunology, School of Medicine, University of California at Davis
 We are very interested in the findings of this study. We specifically want to know if darwinian processes can create new genetic instructions for new functions (new encoded meaning), as is generally assumed. This study does not consider any specific functions, new or not, that the new genes may encode. But with lengthy ORFs and clear evidence of functional constraint, the genes must be doing something. The ones arising from previously noncoding sequences obviously provide something not already expressed in fruitflies. But do the new genes create new meaning?
We are very interested in the findings of this study. We specifically want to know if darwinian processes can create new genetic instructions for new functions (new encoded meaning), as is generally assumed. This study does not consider any specific functions, new or not, that the new genes may encode. But with lengthy ORFs and clear evidence of functional constraint, the genes must be doing something. The ones arising from previously noncoding sequences obviously provide something not already expressed in fruitflies. But do the new genes create new meaning?
 Woodstock of evolution? That's how Nature descibes a meeting among sixteen leading biologists at the Konrad Lorenz Institute for Evolution and Cognition Research in Altenberg, Austria, 10-13 July 2008. The purpose of the meeting was to go beyond the modern synthesis that has held evolutionary theory together for more than sixty years. This is necessary because the existing theory leaves much unexplained —
Woodstock of evolution? That's how Nature descibes a meeting among sixteen leading biologists at the Konrad Lorenz Institute for Evolution and Cognition Research in Altenberg, Austria, 10-13 July 2008. The purpose of the meeting was to go beyond the modern synthesis that has held evolutionary theory together for more than sixty years. This is necessary because the existing theory leaves much unexplained —


 The Trichoplax adhaerens genome contains 98 million base pairs, and an estimated 11,514 protein-coding genes. Unlike other more advanced animals such as fruitflies and nematodes, Trichoplax genes have an intron density (7.6/kb) similar to that of vertebrates (8.5/kb). Many of these introns (82% in conserved regions) occupy the same position and phase as the corresponding ones in humans. Even more interestingly, "Each of the 21 longest gene-rich Trichoplax scaffolds contains segments with a significant concentration of orthologues on one or more human chromosome segments." For example, the illustrated Trichoplax and human segments contain 83 pairs of orthologs, of which the "well-known" ones are connected with red or gray lines (disconnected in our adaptation of this image.)
The Trichoplax adhaerens genome contains 98 million base pairs, and an estimated 11,514 protein-coding genes. Unlike other more advanced animals such as fruitflies and nematodes, Trichoplax genes have an intron density (7.6/kb) similar to that of vertebrates (8.5/kb). Many of these introns (82% in conserved regions) occupy the same position and phase as the corresponding ones in humans. Even more interestingly, "Each of the 21 longest gene-rich Trichoplax scaffolds contains segments with a significant concentration of orthologues on one or more human chromosome segments." For example, the illustrated Trichoplax and human segments contain 83 pairs of orthologs, of which the "well-known" ones are connected with red or gray lines (disconnected in our adaptation of this image.)

 Sponges don't have nerve cells, yet they have genes for directing the formation of nerves. "What was really cool is we took some of these genes and expressed them in frog and flies and the sponge gene became functional – the sponge gene directed the formation of nerves in these more complex animals," says Professor Bernie Degnan (pictured) of the School of Integrative Biology, University of Queensland. His lab is mapping the entire genome of the sea sponge. "We really didn't expect it," he adds.
Sponges don't have nerve cells, yet they have genes for directing the formation of nerves. "What was really cool is we took some of these genes and expressed them in frog and flies and the sponge gene became functional – the sponge gene directed the formation of nerves in these more complex animals," says Professor Bernie Degnan (pictured) of the School of Integrative Biology, University of Queensland. His lab is mapping the entire genome of the sea sponge. "We really didn't expect it," he adds.
 The potential for life on Mars at the Phoenix landing site is believed to be the subject of a forthcoming NASA announcement.
The potential for life on Mars at the Phoenix landing site is believed to be the subject of a forthcoming NASA announcement.